Next-generation gene sequencing has come a long way since the concept was explained in the 1970s. Here, we look at the technology's evolution and the importance of purified water in supporting genomics.
The Evolution of Human Genome Understanding
What makes us human? This fundamental question has driven scientific inquiry for generations. While philosophers might approach this from a theoretical standpoint, the 1970s marked the beginning of a more literal understanding through genetic research. The groundbreaking discovery of DNA's structure by Watson and Crick in the 1950s initiated a five-decade journey, culminating in the complete sequencing of the human genome in 2003.
Foundation of Modern Sequencing: The Sanger Method
The processes that revolutionised genetic research were first outlined in Sanger et al.'s seminal 1977 paper. The Sanger method ingeniously utilised the absence of a hydroxyl group on a 2',3'-dideoxynucleoside triphosphate (ddNTP) to attach a dideoxyribonucleotide to the chain's end. The addition of radioactive α[32P] enabled DNA visualisation through autoradiography, establishing the foundation for modern sequencing techniques.
Next-Generation Sequencing: The Current Landscape
Next-generation sequencing (NGS) represents a quantum leap in DNA analysis capability. These technologies share the ability to parallel sequence millions of DNA templates simultaneously. The evolution from Sanger sequencing involves:
- Clonal amplification of DNA templates on solid support matrices
- Cyclic sequencing methodology
- Replacement of radioactive primers with fluorescent labels
- Advanced detection systems using sophisticated fluorimetry
Modern Applications and Breakthroughs
The impact of NGS extends far beyond basic research:
Clinical Applications:
- Whole Genome Sequencing (WGS)
- Targeted disease research
- Personalised medicine protocols
Emerging Technologies:
- Long-read sequencing platforms
- AI-integrated genomic analysis
- Spatial transcriptomics
- Portable sequencing devices
Critical Infrastructure: The Role of Ultra-Pure Water
While sequencing technology advances, the fundamental need for ultra-pure water remains constant. Modern laboratories require increasingly sophisticated water purification systems to maintain accuracy and reliability in genetic research.
Point-of-Use Solutions
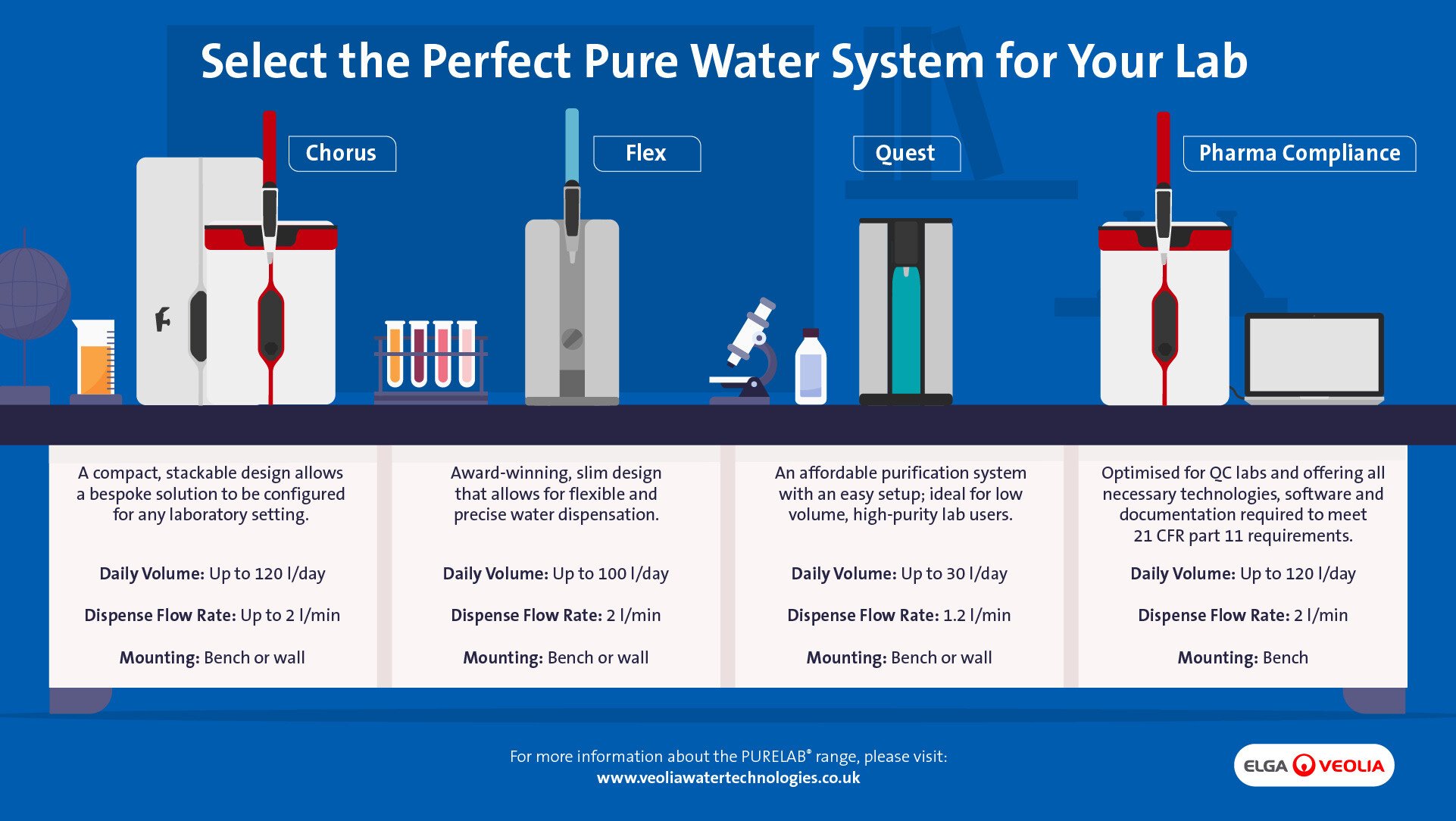
We offer advanced systems, including:
Purelab® Chorus:
- Modular design for flexibility
- Enhanced validation support
- Real-time quality monitoring
- IoT connectivity for remote management
Purelab® Quest:
- Compact footprint for space efficiency
- Three types of water quality output
- Ideal for labs with lower usage requirements
- User-friendly interface
Purelab® Flex 3:
- Significantly reduced carbon footprint through sustainable materials
- Smart water management with advanced RO module
- Eco mode reduces energy consumption by up to 55%
- Superior water purification performance
Centralised Laboratory Systems
- Complete purification, storage and distribution in single unit
- High-volume output up to 30 litres per minute
- Compact design for flexible installation options
- Ideal for new buildings and refurbishments
- Reduced pipework requirements
- Consistent water purity throughout distribution
- Economic operation for large-scale needs
Selecting the Right Water Purification Solution
Choosing appropriate water purification equipment requires careful consideration of:
- Daily volume requirements
- Water quality specifications
- Laboratory layout and infrastructure
- Future scalability needs
For more tips on selecting the right water purification solution, read our blog now. For bespoke support, send a message to one of our experts here.